-
PDF
- Split View
-
Views
-
Cite
Cite
Christoph Varenhorst, Stefan James, David Erlinge, John T. Brandt, Oscar Ö. Braun, Michael Man, Agneta Siegbahn, Joseph Walker, Lars Wallentin, Kenneth J. Winters, Sandra L. Close, Genetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery disease, European Heart Journal, Volume 30, Issue 14, July 2009, Pages 1744–1752, https://doi.org/10.1093/eurheartj/ehp157
Close - Share Icon Share
Abstract
The metabolic pathways leading to the formation of prasugrel and clopidogrel active metabolites differ. We hypothesized that decreased CYP2C19 activity affects the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel.
Ninety-eight patients with coronary artery disease (CAD) taking either clopidogrel 600 mg loading dose (LD)/75 mg maintenance dose (MD) or prasugrel 60 mg LD/10 mg MD were genotyped for variation in six CYP genes. Based on CYP genotype, patients were segregated into two groups: normal function (extensive) metabolizers (EM) and reduced function metabolizers (RM). Plasma active metabolite concentrations were measured at 30 min, 1, 2, 4, and 6 h post-LD and during the MD period on Day 2, Day 14, and Day 29 at 30 min, 1, 2, and 4 h. Vasodilator-stimulated phosphoprotein (VASP) and VerifyNow™ P2Y12 were measured predose, 2, and 24 ± 4 h post-LD and predose during the MD period on Day 14 ± 3 and Day 29 ± 3. For clopidogrel, active metabolite exposure was significantly lower (P = 0.0015) and VASP platelet reactivity index (PRI, %) and VerifyNow™ P2Y12 reaction unit (PRU) values were significantly higher (P < 0.05) in the CYP2C19 RM compared with the EM group. For prasugrel, there was no statistically significant difference in active metabolite exposure or pharmacodynamic response between CYP2C19 EM and RM. Variation in the other five genes demonstrated no statistically significant differences in pharmacokinetic or pharmacodynamic responses.
Variation in the gene encoding CYP2C19 in patients with stable CAD contributes to reduced exposure to clopidogrel's active metabolite and a corresponding reduction in P2Y12 inhibition, but has no significant influence on the response to prasugrel.
Introduction
The standard treatment for patients with acute coronary syndrome includes a combination of dual-antiplatelet therapy, aspirin, and a thienopyridine, which has proven to be efficacious in reducing the rate of recurrent cardiac events including stent thrombosis.1,2 Clopidogrel, the most commonly prescribed thienopyridine, and prasugrel, currently under clinical development, are both orally administered prodrugs that, after absorption, must be converted to an active metabolite.3 The active metabolites inhibit platelet aggregation via antagonism of the P2Y12 receptor.4–6
Recently, we reported greater and faster P2Y12 receptor-mediated platelet inhibition with prasugrel loading dose (LD) 60 mg, maintenance dose (MD) 10 mg compared with clopidogrel 600 mg LD, 75 mg MD owing to a more efficient generation of prasugrel's active metabolite in aspirin-treated patients with stable coronary disease.7 Consistent with the literature, more patients had high residual platelet reactivity following clopidogrel administration than following prasugrel administration.8 Platelet inhibition is closely related to levels of active metabolite formation, and a poor pharmacodynamic response to clopidogrel is most likely primarily caused by differences in absorption or metabolism, or both.9,10
The metabolic pathways for clopidogrel and prasugrel have some key differences. Approximately 85% of a clopidogrel dose is hydrolysed by esterases to an inactive metabolite, which cannot be converted to the active metabolite. The remaining clopidogrel is available to be converted to the active metabolite in a process requiring two sequential cytochrome P450 (CYP)-dependent steps with contribution from CYP3A4/5, CYP2C9, CYP1A2 in one step, and CYP2B6 and CYP2C19 in both steps.11–13 In essence, the esterase pathway competes with the CYP pathway for prodrug, and anything that slows the formation of the active metabolite may ‘shunt’ prodrug to the esterase pathway. Prasugrel, on the other hand, is hydrolysed by esterases into an intermediate precursor of the active metabolite. This intermediate is then oxidized to the active metabolite in a single CYP-dependent step by any one of the four CYP enzymes (with major contributions from CYP3A4/5 and CYP2B6 and minor contributions from CYP2C19 and CYP2C9).14 Formation of the prasugrel active metabolite may thus be mediated by any of the four CYP enzymes, and based on previous studies, they appear to compensate for each other.15
The differences in metabolism suggest that factors which reduce the activity of a CYP enzyme might lead to decreased formation of clopidogrel's active metabolite but would not affect the formation of prasugrel's active metabolite. This hypothesis is supported by the observation that co-administration of ketoconazole, a potent CYP3A4/5 inhibitor, did not affect the overall exposure to prasugrel's active metabolite or the associated pharmacodynamic response, whereas co-administration of ketoconazole with clopidogrel resulted in decreased exposure to clopidogrel's active metabolite and the associated pharmacodynamic response.15
Emerging data suggest that variation in the genes encoding CYP enzymes associated with decreased CYP enzyme activity are associated with an altered pharmacodynamic and, in healthy volunteers, pharmacokinetic response to clopidogrel but not prasugrel.10,16–20 Therefore, we assessed the hypothesis that variation in the function of individual CYP enzymes, especially CYP2C19, would affect generation of the active metabolite and the corresponding pharmacodynamic response under LD/MD conditions in aspirin-treated patients with coronary artery disease (CAD).
Methods
Subjects and study design
The present study was a prespecified part of a randomized, double-blind, double-dummy, two-arm parallel-group study comparing platelet inhibition of prasugrel 60 mg LD/10 mg MD vs. clopidogrel 600 mg LD/75 mg MD conducted in adult male and female patients with stable CAD.7 Subjects were not required to participate in the genetic sample collection to participate in the main study. Therefore, only subjects who signed a specific informed consent document for genotyping were included in these analyses (98 of 110 patients in the primary study). Medications capable of inhibiting CYP enzyme function21 were taken by 17 out of 98 patients (Table 1). The genotyping and associated clinical data were anonymized using a one-way, state-of-the-art encryption algorithm. The study was performed according to ethical principles based on the Declaration of Helsinki and was approved by local ethical review boards.
Concomitant medication capable of inhibiting CYP P450 enzyme function
| CYP inhibited . | Drug . | Prasugrel (n) . | Clopidogrel (n) . |
|---|---|---|---|
| 2C19 | Esomeprazole | 1 | 1 |
| 2C19 | Lansoprazole | 1 | 1 |
| 2C19 | Omeprazole | 3 | 6 |
| 2C19 | Pantoprazole | 0 | 1 |
| 2C9 | Amiodarone | 1 | 1 |
| 3A4/5 | Clarithromycin | 1 | 0 |
| 3A4/5 | Verapamil | 0 | 1 |
| CYP inhibited . | Drug . | Prasugrel (n) . | Clopidogrel (n) . |
|---|---|---|---|
| 2C19 | Esomeprazole | 1 | 1 |
| 2C19 | Lansoprazole | 1 | 1 |
| 2C19 | Omeprazole | 3 | 6 |
| 2C19 | Pantoprazole | 0 | 1 |
| 2C9 | Amiodarone | 1 | 1 |
| 3A4/5 | Clarithromycin | 1 | 0 |
| 3A4/5 | Verapamil | 0 | 1 |
CYP, cytochrome P450; n, number of patients.
Concomitant medication capable of inhibiting CYP P450 enzyme function
| CYP inhibited . | Drug . | Prasugrel (n) . | Clopidogrel (n) . |
|---|---|---|---|
| 2C19 | Esomeprazole | 1 | 1 |
| 2C19 | Lansoprazole | 1 | 1 |
| 2C19 | Omeprazole | 3 | 6 |
| 2C19 | Pantoprazole | 0 | 1 |
| 2C9 | Amiodarone | 1 | 1 |
| 3A4/5 | Clarithromycin | 1 | 0 |
| 3A4/5 | Verapamil | 0 | 1 |
| CYP inhibited . | Drug . | Prasugrel (n) . | Clopidogrel (n) . |
|---|---|---|---|
| 2C19 | Esomeprazole | 1 | 1 |
| 2C19 | Lansoprazole | 1 | 1 |
| 2C19 | Omeprazole | 3 | 6 |
| 2C19 | Pantoprazole | 0 | 1 |
| 2C9 | Amiodarone | 1 | 1 |
| 3A4/5 | Clarithromycin | 1 | 0 |
| 3A4/5 | Verapamil | 0 | 1 |
CYP, cytochrome P450; n, number of patients.
Genetic methodology
DNA isolation and genotyping
DNA was isolated from peripheral blood samples anticoagulated with ethylenediaminetetraacetic acid using the Gentra Puregene DNA Isolation Kit (Qiagen, Hilden, Germany). Genotyping was performed with the Affymetrix Targeted Human Drug Metabolizing Enzyme and Transporter (DMET) 1.0 Assay (Affymetrix, Santa Clara, CA, USA).22 47 genetic variants were measured by the DMET 1.0 Assay, comprising 53 alleles for the six CYP genes analysed (2C19, 2B6, 2C9, 3A5, 3A4, 1A2). One additional allele, not available on the chip, was measured by polymerase chain reaction (PCR)/restriction fragment length polymorphism data (RFLP): CYP2C19*17.23 A total of 48 genetic variants comprising 54 alleles (Table 2) was therefore obtained by combining DMET 1.0 and PCR/RFLP data.
Cytochrome P450 genes
| CYP450 gene . | Star alleles . |
|---|---|
| 2C19 | *1A, *2A, *3, *4, *5A, *6, *7, *8, *9, *10, *12, *13, *14, *17a |
| 2B6 | *1A, *1C, *6, *8, *9, *11, *12, *13, *14, *1 |
| 2C9 | *1A, *2A, *3A, *4, *5, *6, *8, *9, *10, *11A, *12 |
| 3A5 | *1A, *3A, *3B, *3D, *3F, *6, *8, *9, *10 |
| 3A4 | *1A, *17, *18 |
| 1A2 | *1A, *1C, *1D, *1E, *1K, *1L, *7 |
| CYP450 gene . | Star alleles . |
|---|---|
| 2C19 | *1A, *2A, *3, *4, *5A, *6, *7, *8, *9, *10, *12, *13, *14, *17a |
| 2B6 | *1A, *1C, *6, *8, *9, *11, *12, *13, *14, *1 |
| 2C9 | *1A, *2A, *3A, *4, *5, *6, *8, *9, *10, *11A, *12 |
| 3A5 | *1A, *3A, *3B, *3D, *3F, *6, *8, *9, *10 |
| 3A4 | *1A, *17, *18 |
| 1A2 | *1A, *1C, *1D, *1E, *1K, *1L, *7 |
aCYP2C19*17 allele measured by conventional polymerase chain reaction followed by restriction fragment length polymorphism analysis. All remaining alleles genotyped by the Affymetrix Targeted human drug-metabolizing enzymes and transporters (DMET) 1.0 Assay (Affymetrix, Santa Clara, CA, USA).
CYP450, cytochrome P450.
Cytochrome P450 genes
| CYP450 gene . | Star alleles . |
|---|---|
| 2C19 | *1A, *2A, *3, *4, *5A, *6, *7, *8, *9, *10, *12, *13, *14, *17a |
| 2B6 | *1A, *1C, *6, *8, *9, *11, *12, *13, *14, *1 |
| 2C9 | *1A, *2A, *3A, *4, *5, *6, *8, *9, *10, *11A, *12 |
| 3A5 | *1A, *3A, *3B, *3D, *3F, *6, *8, *9, *10 |
| 3A4 | *1A, *17, *18 |
| 1A2 | *1A, *1C, *1D, *1E, *1K, *1L, *7 |
| CYP450 gene . | Star alleles . |
|---|---|
| 2C19 | *1A, *2A, *3, *4, *5A, *6, *7, *8, *9, *10, *12, *13, *14, *17a |
| 2B6 | *1A, *1C, *6, *8, *9, *11, *12, *13, *14, *1 |
| 2C9 | *1A, *2A, *3A, *4, *5, *6, *8, *9, *10, *11A, *12 |
| 3A5 | *1A, *3A, *3B, *3D, *3F, *6, *8, *9, *10 |
| 3A4 | *1A, *17, *18 |
| 1A2 | *1A, *1C, *1D, *1E, *1K, *1L, *7 |
aCYP2C19*17 allele measured by conventional polymerase chain reaction followed by restriction fragment length polymorphism analysis. All remaining alleles genotyped by the Affymetrix Targeted human drug-metabolizing enzymes and transporters (DMET) 1.0 Assay (Affymetrix, Santa Clara, CA, USA).
CYP450, cytochrome P450.
Classification based on predicted metabolic phenotype
To assess the effect of CYP genetic variation on the generation of prasugrel and clopidogrel active metabolite and subsequent pharmacodynamic response, individual variants of six CYP genes known to be involved in the metabolism of the two drugs were classified a priori according to their predicted metabolic phenotypes (normal, increased, or reduced enzymatic function). This classification was defined according to literature-based predictions24,25 using the established common consensus or ‘star allele’ nomenclature (http://www.cypalleles.ki.se).
The combination of two alleles comprises a genotype and the various genotypes (for example, CYP2C19*1A/*1A) for each of the six CYP genes were placed in one of the following categories: extensive metabolizer (EM), defined as two alleles conferring normal or near-normal activity and reduced metabolizer (RM), defined by at least one reduced-function allele. In the case of CYP2C19, individuals with no alleles conferring decreased activity and at least one allele known to enhance activity (CYP2C19*17) were grouped with the EMs. Individuals with an allele conferring decreased activity and a CYP2C19*17 were categorized as unknown. For CYP3A5, individuals with only one allele conferring decreased activity have been found to have near-normal activity24,25 and were therefore categorized as EMs. CYP3A5 RMs were defined as having two reduced function alleles. Table 3 contains a summary of observed genotypes and their corresponding functional categories (predicted phenotypes) used for analyses.
Genotyping results
| Gene . | Predicted phenotype . | Observed genotypes . | Prasugrel, n (%) . | Clopidogrel, n (%) . |
|---|---|---|---|---|
| CYP1A2 | EM | *1A/*1A, *1A/*1D, *1A/*1E, *1D/*1D, *1D/*1E | 49 (96) | 45 (96) |
| RM | *1C/*1D | 0 (0) | 1 (2) | |
| CYP2C19 | EM | *17/*17, *1A/*17, *1A/*1A | 35 (69) | 37 (79) |
| RM | *1A/*2A, *1A/*8, *2A/*2A | 15 (29) | 9 (19) | |
| Uncertain functional status | *2A/*17 | 1 (2) | 1 (2) | |
| CYP2B6 | EM | *1A/*1A, *1A/*1C, *1C/*1C | 29 (57) | 29 (62) |
| RM | *1A/*9, *1C/*9, *9/*9 | 21 (41) | 17 (36) | |
| CYP2C9 | EM | *1A/*1A, *1A/*2A, *1A/*12 | 41 (80) | 40 (85) |
| RM | *1A/*3A, *2A/*2A, *2A/*3A | 9 (18) | 7 (15) | |
| CYP3A4 | EM | *1A/*1A | 51 (100) | 47 (100) |
| CYP3A5 | EM | *1A/*1A, *1A/*3A | 4 (8) | 11 (23) |
| RM | *3A/*3A | 46 (90) | 35 (74) |
| Gene . | Predicted phenotype . | Observed genotypes . | Prasugrel, n (%) . | Clopidogrel, n (%) . |
|---|---|---|---|---|
| CYP1A2 | EM | *1A/*1A, *1A/*1D, *1A/*1E, *1D/*1D, *1D/*1E | 49 (96) | 45 (96) |
| RM | *1C/*1D | 0 (0) | 1 (2) | |
| CYP2C19 | EM | *17/*17, *1A/*17, *1A/*1A | 35 (69) | 37 (79) |
| RM | *1A/*2A, *1A/*8, *2A/*2A | 15 (29) | 9 (19) | |
| Uncertain functional status | *2A/*17 | 1 (2) | 1 (2) | |
| CYP2B6 | EM | *1A/*1A, *1A/*1C, *1C/*1C | 29 (57) | 29 (62) |
| RM | *1A/*9, *1C/*9, *9/*9 | 21 (41) | 17 (36) | |
| CYP2C9 | EM | *1A/*1A, *1A/*2A, *1A/*12 | 41 (80) | 40 (85) |
| RM | *1A/*3A, *2A/*2A, *2A/*3A | 9 (18) | 7 (15) | |
| CYP3A4 | EM | *1A/*1A | 51 (100) | 47 (100) |
| CYP3A5 | EM | *1A/*1A, *1A/*3A | 4 (8) | 11 (23) |
| RM | *3A/*3A | 46 (90) | 35 (74) |
n, Number of subjects; CYP, cytochrome P450; EM, extensive metabolizer; RM, reduced metabolizer.
Genotyping results
| Gene . | Predicted phenotype . | Observed genotypes . | Prasugrel, n (%) . | Clopidogrel, n (%) . |
|---|---|---|---|---|
| CYP1A2 | EM | *1A/*1A, *1A/*1D, *1A/*1E, *1D/*1D, *1D/*1E | 49 (96) | 45 (96) |
| RM | *1C/*1D | 0 (0) | 1 (2) | |
| CYP2C19 | EM | *17/*17, *1A/*17, *1A/*1A | 35 (69) | 37 (79) |
| RM | *1A/*2A, *1A/*8, *2A/*2A | 15 (29) | 9 (19) | |
| Uncertain functional status | *2A/*17 | 1 (2) | 1 (2) | |
| CYP2B6 | EM | *1A/*1A, *1A/*1C, *1C/*1C | 29 (57) | 29 (62) |
| RM | *1A/*9, *1C/*9, *9/*9 | 21 (41) | 17 (36) | |
| CYP2C9 | EM | *1A/*1A, *1A/*2A, *1A/*12 | 41 (80) | 40 (85) |
| RM | *1A/*3A, *2A/*2A, *2A/*3A | 9 (18) | 7 (15) | |
| CYP3A4 | EM | *1A/*1A | 51 (100) | 47 (100) |
| CYP3A5 | EM | *1A/*1A, *1A/*3A | 4 (8) | 11 (23) |
| RM | *3A/*3A | 46 (90) | 35 (74) |
| Gene . | Predicted phenotype . | Observed genotypes . | Prasugrel, n (%) . | Clopidogrel, n (%) . |
|---|---|---|---|---|
| CYP1A2 | EM | *1A/*1A, *1A/*1D, *1A/*1E, *1D/*1D, *1D/*1E | 49 (96) | 45 (96) |
| RM | *1C/*1D | 0 (0) | 1 (2) | |
| CYP2C19 | EM | *17/*17, *1A/*17, *1A/*1A | 35 (69) | 37 (79) |
| RM | *1A/*2A, *1A/*8, *2A/*2A | 15 (29) | 9 (19) | |
| Uncertain functional status | *2A/*17 | 1 (2) | 1 (2) | |
| CYP2B6 | EM | *1A/*1A, *1A/*1C, *1C/*1C | 29 (57) | 29 (62) |
| RM | *1A/*9, *1C/*9, *9/*9 | 21 (41) | 17 (36) | |
| CYP2C9 | EM | *1A/*1A, *1A/*2A, *1A/*12 | 41 (80) | 40 (85) |
| RM | *1A/*3A, *2A/*2A, *2A/*3A | 9 (18) | 7 (15) | |
| CYP3A4 | EM | *1A/*1A | 51 (100) | 47 (100) |
| CYP3A5 | EM | *1A/*1A, *1A/*3A | 4 (8) | 11 (23) |
| RM | *3A/*3A | 46 (90) | 35 (74) |
n, Number of subjects; CYP, cytochrome P450; EM, extensive metabolizer; RM, reduced metabolizer.
Assessment of the active metabolites
Plasma concentrations of the prasugrel active metabolite (R-138727) and clopidogrel active metabolite (R-130964) were analysed in samples obtained at 30 min, 1, 2, 4, and 6 h post-LD and during the MD period on Day 2, Day 14, and Day 29 at 30 min, 1, 2, and 4 h post-MD as previously described.7
Pharmacodynamic assessment of platelet activity
Blood samples were collected into one-tenth volume of 3.2% trisodium citrate from the patients at baseline, 2, and 24 h post-LD and at Day 14 ± 3 and Day 29 ± 3, both before that day's MD. The vasodilator-stimulated phosphoprotein (VASP) assay, a measure of P2Y12 function, was performed using a commercially available method according to the manufacturer's specifications (Biocytex Platelet VASP kit, Marseille, FR) as previously described.7 The platelet reactivity index (PRI, %) was calculated from the corrected mean fluorescence intensity (cMFI) following incubation of the platelets with either prostaglandin E1 alone or prostaglandin E1 + ADP as follows:
The VerifyNow™ P2Y12 assay (VN-P2Y12, Accumetrics, San Diego, CA, USA) is a whole-blood, point-of-care, light transmission-based optical detection assay that measures platelet-induced aggregation in a single-use disposable cartridge containing fibrinogen-coated beads.26 Results from the device are reported as P2Y12 reaction units (PRU) on a continuous scale from 0 upward. The VN-P2Y12 assay was performed on Day 1 at baseline (predose), 2, and 24 ± 4 h post-LD and predose during the MD period on Day 14 ± 3 and Day 29 ± 3.PRI % = [(cMFI(PGE1) − cMFI(PGE1 + ADP))/cMFI(PGE1)] × 100%
Statistical analyses
The primary a priori hypothesis, to evaluate the effect of genetic variation in CYP2C19 on exposure to active metabolite and subsequent platelet aggregation pharmacodynamic responses following treatment with prasugrel or clopidogrel, was investigated. Initially, a linear model testing for interaction between genetic group (EM, RM) and the exposure to active metabolite, the mean log AUC0 −∞ was employed. The log transformation for area under curve (AUC) was used for data normalization. As the interaction model does not specify which drug treatment or genetic group is responsible for the significant effect, if a significant interaction was observed, further comparisons of genetic effect in each of the treatment groups would be undertaken. For the pharmacokinetic analyses, the mean log (AUC0 −∞) of the EM was compared with that of the RM within each treatment group by estimating two contrasts (prasugrel-EM vs. prasugrel-RM and clopidogrel-EM vs. clopidogrel-RM) using a linear model with body weight as a covariate. The statistical significance was assessed via a two-sided test at the 0.05 α level. Prasugrel-EM was also compared with clopidogrel-EM in a similar manner. Analysis was not performed on AUC0 − ∞ at MD since pharmacokinetic parameters were derived from a population-based model that included a component to account for differences between LD and MD.27 For the pharmacodynamic analyses, the mean of the EM group was compared with that of the RM group within each treatment group, and for each pharmacodynamic endpoint [VerifyNow™ (PRU) and VASP (PRI)], by estimating two contrasts (prasugrel-EM vs. prasugrel-RM and clopidogrel-EM vs. clopidogrel-RM) using a similar linear model as in the pharmacokinetic analyses with baseline pharmacodynamic values and body weight as covariates.
Subsequent analyses investigated the contribution of CYP2B6, CYP2C9, CYP1A2, and CYP3A5 to pharmacokinetic and pharmacodynamic responses to either of the thienopyridines. As in the CYP2C19 analysis, the contrasts between EMs and RMs for each gene and within each treatment arm (prasugrel or clopidogrel) were estimated using a linear model with body weight as a covariate.
Results
Patients
Of the 110 patients, 98 participating in the main study consented to genetic testing, 51 in the prasugrel group, and 47 in the clopidogrel group. This genetic subpopulation of patients had similar demographic and clinical characteristics to the overall study population (Table 4).7
Demographics and baseline characteristics
| . | Prasugrel . | Clopidogrel . | ||
|---|---|---|---|---|
| . | TABR genetic subgroup (n = 51) . | TABR entire study (n = 55) . | TABR genetic subgroup (n = 47) . | TABR entire study (n = 55) . |
| Age (mean ± SD) | 62.6 ± 6.1 | 62.0 ± 6.1 | 65.0 ± 5.7 | 64.0 ± 6.2 |
| Body weight (mean ± SD) | 88.4 ± 12.8 | 87.3 ± 13.5 | 85.6 ± 11.9 | 84.3 ± 11.7 |
| Gender, n (%) | ||||
| Female | 7 (14) | 7 | 2 (4) | 2 |
| Male | 44 (86) | 48 | 45 (96) | 53 |
| Smoking status, n (%) | ||||
| No | 43 (84) | 46 | 43 (91) | 50 |
| Yes | 8 (16) | 9 | 4 (9) | 5 |
| Diabetes, n (%) | ||||
| No | 40 (78) | 44 (80) | 39 (83) | 46 (84) |
| Yes | 11 (22) | 11 (20) | 8 (17) | 9 (16) |
| Ethnicity, n (%) | ||||
| Caucasian | 51 (100) | 55 (100) | 47 (100) | 55 (100) |
| . | Prasugrel . | Clopidogrel . | ||
|---|---|---|---|---|
| . | TABR genetic subgroup (n = 51) . | TABR entire study (n = 55) . | TABR genetic subgroup (n = 47) . | TABR entire study (n = 55) . |
| Age (mean ± SD) | 62.6 ± 6.1 | 62.0 ± 6.1 | 65.0 ± 5.7 | 64.0 ± 6.2 |
| Body weight (mean ± SD) | 88.4 ± 12.8 | 87.3 ± 13.5 | 85.6 ± 11.9 | 84.3 ± 11.7 |
| Gender, n (%) | ||||
| Female | 7 (14) | 7 | 2 (4) | 2 |
| Male | 44 (86) | 48 | 45 (96) | 53 |
| Smoking status, n (%) | ||||
| No | 43 (84) | 46 | 43 (91) | 50 |
| Yes | 8 (16) | 9 | 4 (9) | 5 |
| Diabetes, n (%) | ||||
| No | 40 (78) | 44 (80) | 39 (83) | 46 (84) |
| Yes | 11 (22) | 11 (20) | 8 (17) | 9 (16) |
| Ethnicity, n (%) | ||||
| Caucasian | 51 (100) | 55 (100) | 47 (100) | 55 (100) |
n, number of patients; SD, standard deviation. Body weight is measured in kilograms, age is measured years. TABR Genetic Subgroup: those patients in TABR who provided a sample for genetics.
Demographics and baseline characteristics
| . | Prasugrel . | Clopidogrel . | ||
|---|---|---|---|---|
| . | TABR genetic subgroup (n = 51) . | TABR entire study (n = 55) . | TABR genetic subgroup (n = 47) . | TABR entire study (n = 55) . |
| Age (mean ± SD) | 62.6 ± 6.1 | 62.0 ± 6.1 | 65.0 ± 5.7 | 64.0 ± 6.2 |
| Body weight (mean ± SD) | 88.4 ± 12.8 | 87.3 ± 13.5 | 85.6 ± 11.9 | 84.3 ± 11.7 |
| Gender, n (%) | ||||
| Female | 7 (14) | 7 | 2 (4) | 2 |
| Male | 44 (86) | 48 | 45 (96) | 53 |
| Smoking status, n (%) | ||||
| No | 43 (84) | 46 | 43 (91) | 50 |
| Yes | 8 (16) | 9 | 4 (9) | 5 |
| Diabetes, n (%) | ||||
| No | 40 (78) | 44 (80) | 39 (83) | 46 (84) |
| Yes | 11 (22) | 11 (20) | 8 (17) | 9 (16) |
| Ethnicity, n (%) | ||||
| Caucasian | 51 (100) | 55 (100) | 47 (100) | 55 (100) |
| . | Prasugrel . | Clopidogrel . | ||
|---|---|---|---|---|
| . | TABR genetic subgroup (n = 51) . | TABR entire study (n = 55) . | TABR genetic subgroup (n = 47) . | TABR entire study (n = 55) . |
| Age (mean ± SD) | 62.6 ± 6.1 | 62.0 ± 6.1 | 65.0 ± 5.7 | 64.0 ± 6.2 |
| Body weight (mean ± SD) | 88.4 ± 12.8 | 87.3 ± 13.5 | 85.6 ± 11.9 | 84.3 ± 11.7 |
| Gender, n (%) | ||||
| Female | 7 (14) | 7 | 2 (4) | 2 |
| Male | 44 (86) | 48 | 45 (96) | 53 |
| Smoking status, n (%) | ||||
| No | 43 (84) | 46 | 43 (91) | 50 |
| Yes | 8 (16) | 9 | 4 (9) | 5 |
| Diabetes, n (%) | ||||
| No | 40 (78) | 44 (80) | 39 (83) | 46 (84) |
| Yes | 11 (22) | 11 (20) | 8 (17) | 9 (16) |
| Ethnicity, n (%) | ||||
| Caucasian | 51 (100) | 55 (100) | 47 (100) | 55 (100) |
n, number of patients; SD, standard deviation. Body weight is measured in kilograms, age is measured years. TABR Genetic Subgroup: those patients in TABR who provided a sample for genetics.
Genotyping results
The overall genotyping success rate was 98.8% with <1.2% genotypes unable to be called (Table 3). CYP2C19, CYP2C9, and CYP3A4 had a 100% genotyping success rate with all alleles determined for all patients for these genes.
The frequencies of predicted metabolic phenotypes were similar to the published Caucasian frequencies (Table 4; see http://www.cypalleles.ki.se). The frequency of RM was sufficient to complete the analyses for CYP3A5, CYP2C19, CYP2C9, and CYP2B6. However, the observed frequency of RM for CYP3A4 and CYP1A2 was too low to support the statistical analyses with no RM patients for CYP3A4 and only one RM patient observed for CYP1A2.
Relationship between pharmacokinetics and CYP2C19
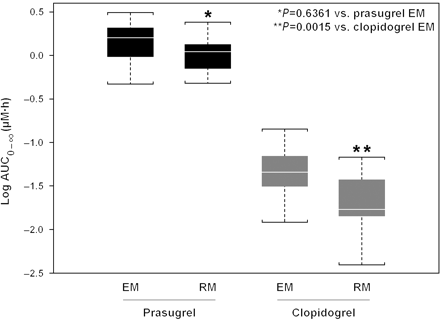
CYP2C19 EM/RM and PK interaction was evaluated first using linear model and was found statistically significant. In prasugrel-treated patients, there was no relationship between exposure to its active metabolite and CYP2C19-predicted metabolizer status (P = 0.6361). Conversely, in clopidogrel-treated patients, a statistically significant lower total plasma exposure (AUC) of clopidogrel active metabolite was observed in RMs compared to those with CYP2C19 EM status (P = 0.0015; Figures 1 and 2). Exposure for prasugrel active metabolite was higher for the prasugrel EM group compared with the clopidogrel EM group (P = 0.000). That is, the active metabolite exposure following the prasugrel 60 mg LD was higher than after the clopidogrel 600 mg LD even when the comparison was limited to patients with normal CYP2C19 activity as predicted by genotype.
Comparison of prasugrel 60 mg and clopidogrel 600 mg loading dose exposure of active metabolite by CYP2C19 genetic classification. Box represents median, 25th, and 75th percentiles and whiskers represent the most extreme values within 1.5 times inter-quartile range of the box. AUC, area under the concentration–time curve; EM, extensive metabolizer; RM, reduced metabolizer.
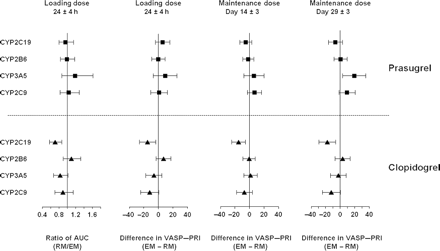
Ratio RM/EM or difference (EM − RM) for pharmacokinetics and pharmacodynamic responses for CYP2C19, CYP2B6, CYP2C9, and CYP3A5. Mean and 95% confidence interval for ratio (AUC at LD) or difference (VASP–PRI at 24 h post-LD and MD Day 14 and Day 29) is derived from a linear model and is plotted for each CYP gene. AUC, area under the concentration–time curve; EM, extensive metabolizer; LD, loading dose; MD, maintenance dose; RM, reduced metabolizer; VASP, vasodilator-stimulated phosphoprotein; PRI, platelet reactivity index.
Relationship between pharmacodynamic response and CYP2C19
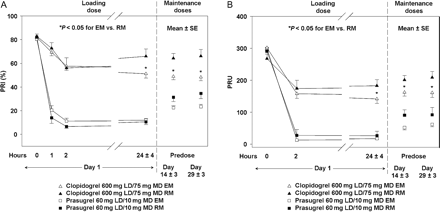
In clopidogrel-treated patients, patients with CYP2C19 RM genotypes exhibited a reduced pharmacodynamic response, as measured by VASP, compared with those with CYP2C19 EM genotypes (Figure 3A). Similar results were seen with the point-of-care device VerifyNow™ P2Y12 (Figure 3B). This difference was not seen for either the VASP assay or the VerifyNow™ P2Y12 device in CYP2C19 EM and RM in patients treated with prasugrel.
Relationship between pharmacodynamic responses and CYP2C19 genetic classification measured by VASP assay (A) and VerifyNow™ P2Y12 device (B). EM, extensive metabolizer; RM, reduced metabolizer; VASP, vasodilator-stimulated phosphoprotein; PRI, platelet reactivity index; PRU, P2Y12 reaction unit.
Relationship between pharmacokinetics/pharmacodynamics and other analysed CYP P450 genes
For CYP2C9, CYP2B6, and CYP3A5, there was no statistically significant effect of genotype on the exposure to active metabolite exposure for either drug (Figure 2). Accordingly, as expected, given the strong relationship between levels of active metabolite and degree of P2Y12 inhibition, analyses on their pharmacodynamic responses illustrated no statistically significant effect on PRI measured by VASP assay (Figure 2) or VerifyNow™ P2Y12 device.
Discussion
The results of this study support the hypothesis that a decrease in CYP2C19 function reduces the formation of clopidogrel's active metabolite, resulting in less inhibition of platelet function. In contrast, decreased CYP2C19 function does not appear to affect the exposure to prasugrel's active metabolite or the resulting inhibition of platelet function. These findings in a cohort of aspirin-treated patients with CAD confirm the previously described observations in healthy subjects.10,12 Of note, this genetic effect was even evident following the higher, 600 mg clopidogrel LD and persisted throughout the 75 mg MD phase. These results demonstrate, in a patient cohort, that genetic variation resulting in decreased CYP function partly accounts for poor pharmacodynamic response to clopidogrel and lower exposure to clopidogrel active metabolite. Our findings confirm the critical link between exposure to active metabolite and subsequent platelet response in a patient population. Our data also agree well with results reported by others where the CYP2C19*2 allele was associated with a higher on-clopidogrel platelet reactivity which in turn has been linked to worsened clinical outcome after coronary stenting.16,19,28,29 The effect of variation in CYP2C19 on responsiveness to clopidogrel complicates the prescribing of thienopyridines.
The frequency of CYP2C19 RM genotypes shows wide inter-ethnic variation, ranging from 20–30% in Caucasians to 35–45% in African-Americans and 50–65% in East Asians. The most common defective allele, CYP2C19*2, accounts for 75–85% of the CYP2C19 alleles responsible for RMs in Caucasians and East Asians.30 The ethnic variation in CYP2C19*2 is likely one of the causes for inter-ethnic differences in the pharmacokinetics of several widely prescribed drugs that are substrates for CYP2C19 and also suggests that the response to clopidogrel may vary by ethnicity.18
In the present trial, the results were potentially confounded by the nearly 20% of patients (17 of 98) who were taking medications known to inhibit CYP function (14 of these patients were taking CYP2C19 inhibitors such as omeprazole and lansoprazole). In a post hoc analysis, patients receiving medication purported to be CYP2C19 inhibitors were grouped together with the CYP2C19 RM genetic group. The difference in pharmacokinetic and pharmacodynamic response to clopidogrel between the modified EM and RM groups was even more apparent (data not shown). For prasugrel, no such trend was observed when patients receiving potential CYP2C19-inhibitor drugs were grouped with the CYP2C19 RM group. In addition to inhibition of CYP2C19, another possible mechanism for drug-interaction with proton pump inhibitors (PPI) is alteration in gastric pH, which could affect thienopyridine absorption. In a recent cross-over study, increasing gastric pH with lansoprazole did not decrease the level of platelet inhibition after a prasugrel 60 mg LD, while in contrast in the same subjects lansoprazole did tend to dampen the antiplatelet response observed with a clopidogrel 300 mg LD.9
Both clopidogrel- and prasugrel-active metabolites show similar antiplatelet activity and exposure–pharmacodynamic relationships.27,31 We have previously reported a strong correlation between plasma concentrations of active metabolite and platelet inhibition for both prasugrel and clopidogrel up to saturation levels of the P2Y12 receptor.7 Addition of clopidogrel active metabolite ex vivo resulted in maximal platelet inhibition, even in subjects previously classified as clopidogrel poor responders.7 Although polymorphisms of the P2Y12 receptor have been associated with different degrees of platelet aggregation in healthy volunteers, the P2Y12 H2 haplotype has not been proven to modulate clopidogrel response in patients with CAD.29,32–34 In addition, as the P2Y12 receptor is the target for both prasugrel and clopidogrel and the active metabolite of prasugrel and clopidogrel have been shown to be nearly equipotent in vitro, P2Y12 receptor variants would likely have similar effects on the response to both drugs.35 Together, these data strongly suggest that a poor pharmacodynamic response to clopidogrel is caused by ineffective generation of the active metabolite rather than P2Y12 receptor heterogeneity.7
For clopidogrel, only a fraction of the prodrug is converted to the active metabolite in two sequential CYP-dependent steps. For prasugrel, the prodrug is rapidly converted first to an intermediate by esterases and then in one single CYP-dependent step to the active metabolite. Consequently, variation in the function of individual CYP-enzymes is not likely to affect the generation of the prasugrel active metabolite since any of the several CYPs can mediate the oxidation of the intermediate thiolactone metabolite.14 For clopidogrel on the other hand, with approximately 85% of the prodrug converted to the inactive metabolite, even small changes on CYP enzyme activity seem to influence formation of its active metabolite.36
Changes in CYP enzyme activity are known to be related to both genetic variants that result in decreased CYP function and the use of concomitant CYP-inhibitory drugs. For example, as previously reported, patients randomized to clopidogrel and aspirin co-administered with omeprazole, a drug capable of inhibiting CYP2C19, had a significantly decreased platelet inhibitory effect, and the risk of being classified as a poor responder was 4.31 times greater compared with patients treated only with clopidogrel.37 These results are consistent with the recently presented findings of the MedCo study in which patients taking both clopidogrel and PPIs experienced a higher rate of cardiac events than those taking clopidogrel alone.38
When examining other CYPs, no statistically significant association was found between genetic variants and the response to thienopyridines, clopidogrel, or prasugrel. The effect on clopidogrel's antiplatelet response by CYP3A substrates, such as lipophilic statins, has been debated, and the results reported are contradictory.39–41 In a cross-over study on healthy subjects, ketoconazole, a potent inhibitor of CYP3A4/3A5 and representing a ‘worst-case’ interaction for CYP3A substrates, was shown to decrease the exposure to the active metabolite of clopidogrel but not prasugrel.15 As the clinical implication remains uncertain, further investigation on interactions of lipophilic statins with thienopyridine treatment is warranted. CYP2C9 loss-of-function genotypes have been associated with decreased exposure to the active metabolite for clopidogrel but not prasugrel; however, this observation was not confirmed in this patient population.10
Study limitations
Some limitations in this study need to be addressed. Because of sample size we did not observe sufficient numbers of patients with decreased function polymorphisms for CYP3A4 and CYP1A2 to evaluate a possible relationship to decreased thienopyridine response. Nonetheless, decreased function polymorphisms of these CYPs are uncommon. CYP1A2 is not involved in prasugrel metabolism. The CYP3A system is known to be involved in the metabolism of both drugs, and inhibition has been demonstrated to result in decreased exposure to clopidogrel.15 The role of individual CYP3A genes in thienopyridine activation is complicated by the known compensatory actions between CYP3A4 and CYP3A5. Furthermore, the low frequency of variants in all ethnic subgroups for CYP3A4 and the high degree of null variants in CYP3A5 do not directly correlate with the level of clopidogrel unresponsiveness. Thus, variants in these genes are unlikely to play a clinically meaningful role in variable response to clopidogrel or prasugrel. However, although variants with established functional effect were measured, further investigation is warranted as not all variants for CYP3A4 were tested in the current study.
We grouped genotypes characterized as moderately reduced and those with essentially no reported activity into a single group, RM. Although essential for power, we therefore could not assess whether there was a ‘gene dose–effect’ for either the response to prasugrel or clopidogrel.18,20 A larger study population would allow for further discrimination between those with moderately reduced and those with ablated CYP2C19 function. In addition, as subjects in this study were not followed long-term to collect clinical outcome measures, it was not possible to establish a relationship between variation in the genes encoding CYP enzymes and clinical outcome. However, the loss-of-function CYP2C19*2 allele has been associated with high on-clopidogrel platelet reactivity, a phenotype that has been linked to a poorer clinical outcome.16,19,28 This observation has been re-inforced by three recent publications reporting that reduced function CYP2C19 genotypes were associated with an increased rate of clinical events in patients with acute coronary syndrome.19,28,29
Despite the above stated limitations, this study represents, to our knowledge, the most comprehensive genotyping assessment ever completed on a patient population exposed to thienopyridines. Thus, this is the first time a comprehensive assay, assessing all the variation predicted to result in decreased function, in six of the genes involved in the metabolism of thienopyridines has been investigated. In addition to the uniqueness of the genotyping, this is the first time that variation in genes encoding CYP enzymes conferring reduced function has been directly linked to reduced pharmacokinetic exposure and also to reduced pharmacodynamic response in a patient population.
This study showed that variation in the gene encoding CYP2C19 contributes to reduced exposure to clopidogrel's active metabolite and a corresponding reduction in P2Y12 inhibition, but has no influence on the response to prasugrel. The usefulness of assessment of this and other genetic polymorphisms for the selection of type and dose of thienopyridine needs further evaluation in clinical outcome trials.
Funding
Daiichi Sankyo Company Limited and Eli Lilly and Company (to Drs C.V., S.J., D.E., O.Ö.B., L.W., and A.S.). Funding to pay the Open Access publication charges for this article was provided by Daiichi Sankyo Company, Limited, Tokyo, Japan and Eli Lilly and Company, Indianapolis, IN, USA.
Conflict of interest: J.T.B., S.L.C., M.M., and K.J.W. are employees and stockholders of Eli Lilly and Company. J.W. is an employee of Daiichi Sankyo Inc.
Acknowledgements
The authors would like to thank Vivian Thieu for writing and administrative assistance (Eli Lilly and Company) and Julie Sherman for editorial assistance.